3d visualization with VMD
Visual Molecular Dynamics can display, animate, and analyze large biomolecular systems using 3-D graphics and built-in scripting.
Again, we are going to use only a small subset of VMD's powers here. You are very welcome to dig deeper in the nice VMD tutorial.
Starting VMD
vmd # start vmd vmd pos.xyz # start vmd and load position file 'pos.xyz' vmd -e view.vmd # start vmd and load previously saved visualization state 'view.vmd'
VMD does have a graphical user interface (yay!), although knowing how to use the scripting console is also advisable. Some common tasks are:
- Edit representation of structure: Graphics → Representations
- Measure distance between atoms: Mouse → Label → Bonds
- Plot a specific bond length versus frame number: Graphics → Labels → Bonds → Graph
- Render the current view as bitmap image (
.bmp): File → Render.. → Snapshot. - Note: Click 'Browse' to place the
.bmpin the desired directory.
We are going to start by creating a visual representation of the protein rubredoxin.
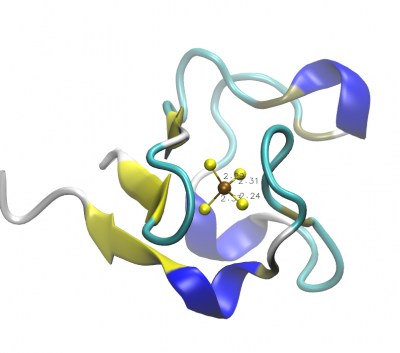
- From the RCSB Protein Data Bank download the PDB file for rubredoxin. We will use the structure determined by Watenpaugh et al. with PDB ID
4RXN. - Visualize the protein with VMD. Suggestion: Draw the backbone with NewCartoon and color it according to the secondary structure.
- Find the $\text{FeS}_4$ active site and measure the 4 $\text{Fe}-\text{S}$ distances.
- Render a snapshot of the active site with the measured distances.
Now we are ready to analyze a molecular dynamics trajectory.
The file nacl.xyz in the intro folder contains the trajectory of a MD simulation of $\text{NaCl}$ in water, which naturally was performed under periodic boundary conditions.
For simulations with periodic boundary conditions it is often helpful to draw the simulation box. In VMD this is achieved by typing the following (for a cubic simulation cell with 10 angstroms side length) on the terminal
pbc set {10 10 10} -all
pbc box
- How many water molecules were simulated?
- Visualize the trajectory with VMD. You may want to change the representation style from 'lines' to something else.
- Measure the $\text{Na-Cl}$ distance and create a plot of its evolution with the frame number.
- The simulation was performed in a cubic box with side length 12.4138 Angstroms and periodic boundary conditions. Draw the simulation box.
- In the course of the simulation, the molecules start moving outside of the box! Does this indicate a problem?
- Compare with the premade visualization state
nacl.vmd. Describe the mathematical transformation that has been applied here.
