Table of Contents
GGA based surface science
GGA DFT calculations can be performed with CP2K relatively easily for many systems. While the CP2K reference manual lists all of the various options, the initial setup of a system is easy, and few details about the internals needs to be known. The input files provide here form a good template to start.
Crucial to start are a reasonable initial structure, and for condensed phase systems the size of the simulation cell. Once these are known, copy&paste might be enough to start the simulation.
Among the important parameters of the simulations are:
- model (structure)
- Gaussian basis set
- Plane Waves (PW) cutoff
- Density functional
Assessing the influence of these parameters might be more challenging.
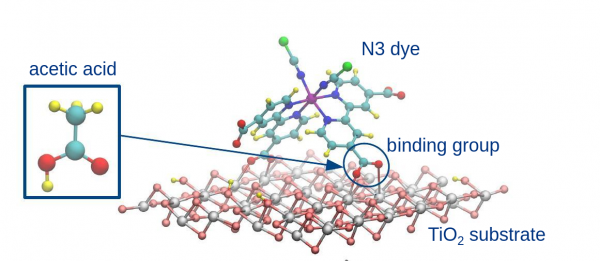
Dye anchoring to TiO$_2$
In this exercise you will compare two possible binding modes of acetic acid to anatase TiO$_2$. Acetic acid contains the carboxylic group. It is commonly used in Dye-Sensitized Solar Cells as an anchoring moiety to bind light harvesting dyes to semi-conducting substrates. We will therefore use acetic acid as a model of the more complex dye molecules, as done in this paper: 10.1021/jp4117563. To speedup calculations, only the smallest slab model is employed.
1. Task: Familiarize yourself with system and setup
- Use vmd to vizualize the geometries (provided below) named
mode1.xyzandmode2.xyz - To edit the input files provided below, use an editor such as
viornano. Whilenanois simple to use,vican be configured to colour-code cp2k inputs. - You will need files named
BASIS_MOLOPTGTH_POTENTIALSdftd3.datthat are provided as part of CP2K in a directorycp2k/data, unless the code has been compiled with the proper flag (-D__DATA_DIR) so that these are found automatically. - Use a job script to submit jobs on the cluster, an example job submission script might look like
- job
#PBS -N mode1 #PBS -j oe #PBS -q dist_small #PBS -l nodes=4:ppn=16 #PBS -l walltime=10:00 #PBS -A cp2k2015 cd $PBS_O_WORKDIR module purge module load cp2k/2.6 prun cp2k.popt -i mode1.inp -o mode1.out
2. Task: Binding induced density differences
We start with single point energy calculations on binding mode 1, to visualize the interaction between molecule and surface. The goal is to compute the binding induced density difference:
\[ \rho_\text{induced}= \rho_\text{slab-dye-complex} - \rho_\text{dye} - \rho_\text{slab} \]
First, we'll discuss in detail the structure and the choices made in the sample input file mode1.inp.
topics:
- Project name
- Runtype
- Gaussian Basis, pseudopotentials
- PW Cutoff
- thresholds
- SCF: OT
- XC and -D3 correction
- Unit cell choice
Second, we run the cp2k input and store the output for analysis and discussion.
cp2k.popt -i mode1.inp -o mode1.out
In addition to the output mode1.out, other files are gerenated by CP2K named MODE1*
topics:
- General overiew
- OT output
- Various grid quantities
- Density cube output
- Timing report
Third, we compute the changes in density induced by the binding. For this you will have to run three separate energy calculations:
- combined system bound in the first mode (file
mode1.xyz) - lone acetic acid molecule (just remove slab's coordinates from
mode1.xyz), name the filemode1_dye.xyz - lone TiO$_2$ slab (just remove the acid's coordinates from
mode1.xyz), name the filemode1_slab.xyz
Create the .xyz files (check with vmd that they contain the right subsystems), and create mode1_dye.inp, mode1_slab.inp by changing both COORD_FILE_NAME and PROJECT accordingly.
After computing these input files, we analyze the results using a tool provided with cp2k cubecruncher.x that manipulates cube files (e.g. can compute difference).
~$ cubecruncher.x -i MODE1-ELECTRON_DENSITY-1_0.cube -subtract MODE1_dye-ELECTRON_DENSITY-1_0.cube -o tmp.cube ~$ cubecruncher.x -i tmp.cube -subtract MODE1_slab-ELECTRON_DENSITY-1_0.cube -center geo -o MODE1_delta.cube
You can visualize the resulting file MODE1_delta.cube with VMD.
3. Task: relative stabilities
In order to compute the relative stability of mode1 and mode2, both configurations need to be geometry optimized.
To do so, turn off the generation of cubes (&E_DENSITY_CUBE OFF) in mode1.inp, change to RUN_TYPE GEO_OPT and adjust the project name. Create and run a similar input file for mode2.
input topics:
- BFGS vs LBFGS
- EPS_SCF, CUTOFF, MAX_DR, ..
output topics:
Informations at step- Trajectory
MODE1_GEO-pos-1.xyz
Compare the final energies (ENERGY| Total FORCE_EVAL ( QS ) energy (a.u.):), and determine which mode is most stable. Does this agree with the values in table 1 of the manuscript cited ?
4. Task: ab initio molecular dynamics
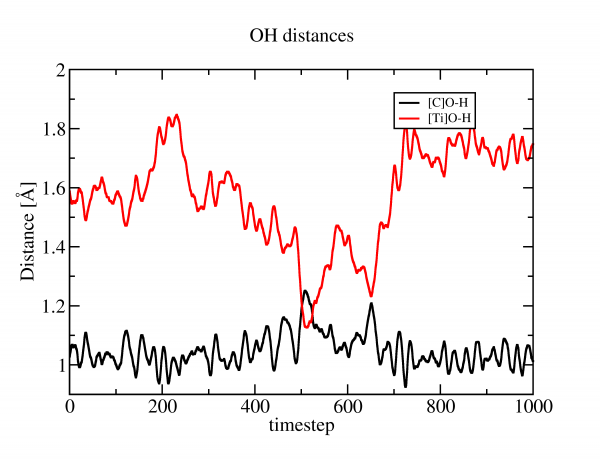
Perform a short ab initio molecular dynamics simulation of the system (~1000 steps, ~0.5ps) by changing to RUN_TYPE MD. After a couple of hours the job should be finished. Now analyze the OH distance in VMD. A possible outcome is :
What can you say about the hydrogen bond to the surface, relative acidity of the two oxygens ? Note that, in order to be statistically relevant, longer trajectories should be employed, and surface slab thickness will play an important role. Also compare to Fig. 7 of the paper referenced.
Required Files
(right) click on the filename to download to your local machine.
- mode1.inp
&GLOBAL ! the project name is made part of most output files... useful to keep order PROJECT MODE1 ! various runtypes (energy, geo_opt, etc.) available. RUN_TYPE ENERGY &END GLOBAL &FORCE_EVAL ! the electronic structure part of CP2K is named Quickstep METHOD Quickstep &DFT ! basis sets and pseudopotential files can be found in cp2k/data BASIS_SET_FILE_NAME BASIS_MOLOPT POTENTIAL_FILE_NAME GTH_POTENTIALS ! Charge and multiplicity CHARGE 0 MULTIPLICITY 1 &MGRID ! PW cutoff ... depends on the element (basis) too small cutoffs lead to the eggbox effect. ! certain calculations (e.g. geometry optimization, vibrational frequencies, ! NPT and cell optimizations, need higher cutoffs) CUTOFF [Ry] 400 &END &QS ! use the GPW method (i.e. pseudopotential based calculations with the Gaussian and Plane Waves scheme). METHOD GPW ! default threshold for numerics ~ roughly numerical accuracy of the total energy per electron, ! sets reasonable values for all other thresholds. EPS_DEFAULT 1.0E-10 ! used for MD, the method used to generate the initial guess. EXTRAPOLATION ASPC &END &POISSON PERIODIC XYZ ! the default, gas phase systems should have 'NONE' and a wavelet solver &END &PRINT ! at the end of the SCF procedure generate cube files of the density &E_DENSITY_CUBE ON &END E_DENSITY_CUBE &END ! use the OT METHOD for robust and efficient SCF, suitable for all non-metallic systems. &SCF SCF_GUESS ATOMIC ! can be used to RESTART an interrupted calculation MAX_SCF 30 EPS_SCF 1.0E-6 ! accuracy of the SCF procedure typically 1.0E-6 - 1.0E-7 &OT ! an accurate preconditioner suitable also for larger systems PRECONDITIONER FULL_SINGLE_INVERSE ! the most robust choice (DIIS might sometimes be faster, but not as stable). MINIMIZER CG &END OT &OUTER_SCF ! repeat the inner SCF cycle 10 times MAX_SCF 10 EPS_SCF 1.0E-6 ! must match the above &END &END SCF ! specify the exchange and correlation treatment &XC ! use a PBE functional &XC_FUNCTIONAL &PBE &END &END XC_FUNCTIONAL ! adding Grimme's D3 correction (by default without C9 terms) &VDW_POTENTIAL POTENTIAL_TYPE PAIR_POTENTIAL &PAIR_POTENTIAL PARAMETER_FILE_NAME dftd3.dat TYPE DFTD3 REFERENCE_FUNCTIONAL PBE R_CUTOFF [angstrom] 16 &END &END VDW_POTENTIAL &END XC &END DFT ! description of the system &SUBSYS &CELL ! unit cells that are orthorhombic are more efficient with CP2K ABC [angstrom] 10.2270 11.3460 20.000 &END CELL ! atom coordinates can be in the &COORD section, ! or provided as an external file. &TOPOLOGY COORD_FILE_NAME mode1.xyz COORD_FILE_FORMAT XYZ &END ! MOLOPT basis sets are fairly costly, ! but in the 'DZVP-MOLOPT-SR-GTH' available for all elements ! their contracted nature makes them suitable ! for condensed and gas phase systems alike. &KIND H BASIS_SET DZVP-MOLOPT-SR-GTH POTENTIAL GTH-PBE-q1 &END KIND &KIND C BASIS_SET DZVP-MOLOPT-SR-GTH POTENTIAL GTH-PBE-q4 &END KIND &KIND O BASIS_SET DZVP-MOLOPT-SR-GTH POTENTIAL GTH-PBE-q6 &END KIND &KIND Ti BASIS_SET DZVP-MOLOPT-SR-GTH POTENTIAL GTH-PBE-q12 &END KIND &END SUBSYS &END FORCE_EVAL ! how to propagate the system, selection via RUN_TYPE in the &GLOBAL section &MOTION &GEO_OPT OPTIMIZER BFGS ! Good choice for 'small' systems (use LBFGS for large systems) MAX_ITER 100 MAX_DR [bohr] 0.003 ! adjust target as needed &BFGS &END &END &MD ENSEMBLE NVT ! sampling the canonical ensemble, accurate properties might need NVE TEMPERATURE [K] 300 TIMESTEP [fs] 0.5 STEPS 1000 # GLE thermostat as generated at http://epfl-cosmo.github.io/gle4md # GLE provides an effective NVT sampling. &THERMOSTAT REGION MASSIVE TYPE GLE &GLE NDIM 5 A_SCALE [ps^-1] 1.00 A_LIST 1.859575861256e+2 2.726385349840e-1 1.152610045461e+1 -3.641457826260e+1 2.317337581602e+2 A_LIST -2.780952471206e-1 8.595159180871e-5 7.218904801765e-1 -1.984453934386e-1 4.240925758342e-1 A_LIST -1.482580813121e+1 -7.218904801765e-1 1.359090212128e+0 5.149889628035e+0 -9.994926845099e+0 A_LIST -1.037218912688e+1 1.984453934386e-1 -5.149889628035e+0 2.666191089117e+1 1.150771549531e+1 A_LIST 2.180134636042e+2 -4.240925758342e-1 9.994926845099e+0 -1.150771549531e+1 3.095839456559e+2 &END GLE &END THERMOSTAT &END &END
- mode1.xyz
116 Ti -0.0179479198 -0.0078042700 -0.0922850421 O 0.4497915479 0.0007412647 1.9321232563 Ti -1.4316444492 -0.0089893346 2.8221490912 O -1.7887110897 0.0007867236 0.7998892697 O -3.2081345298 0.0411605531 3.6695321087 O -1.8010755493 -1.8874663742 2.8216777686 Ti -3.7269830403 -1.9006613639 3.6517557753 O 0.3830863054 -1.8910765741 -0.0700021876 Ti 3.7557586944 -1.8917451634 2.9143676586 O 3.3146470921 -0.0011081937 2.8729289358 O -4.6680663094 -1.8855446073 2.0338712494 O -5.4086768695 -1.8843459443 4.3991860126 Ti -0.0282285127 -3.7757033787 -0.0787156736 O 0.4462623263 -3.7813137083 1.9359917441 Ti -1.3999537875 -3.7707056149 2.8874421519 O -1.7903153040 -3.7850263943 0.8021001791 O -3.2390175951 -3.7978185279 3.6716494047 O -1.7864035184 -5.6734792481 2.8627360747 Ti -3.7357623244 -5.6788609282 3.4482649850 O 0.3837694100 -5.6723382681 -0.0690470865 Ti 3.7205660980 -5.6668034549 2.9114784222 O 3.3236114384 -3.7787921003 2.8721776992 O -4.6689995455 -5.6807480198 1.9358553242 O -5.3525041570 -5.6742752842 4.3359372112 Ti -0.0314402214 3.7828933451 -0.0777664638 O 0.4518754735 3.7839662632 1.9399715530 Ti -1.3792280628 3.7857276911 2.9025413153 O -1.7909627210 3.7866794195 0.8027290293 O -3.2026665631 3.7827561658 3.7306777258 O -1.7857471998 1.9013100770 2.8154049594 Ti -3.7307612355 1.9120465523 3.4387577457 O 0.3836352141 1.8909325251 -0.0691199251 Ti 3.7223424203 1.8887162901 2.9133081650 O 3.3250830331 3.7807500488 2.8687333178 O -4.6728325926 1.9008997868 1.9326739591 O -5.3457700344 1.8899459254 4.3344454640 Ti -1.4852085406 -0.0011927086 -3.7100466053 O -1.0850586016 -0.0008795951 -1.6147031707 Ti -2.8526945884 -0.0039467894 -0.7412906371 O -3.3269327110 -0.0004491456 -2.7420242486 O -4.7289708978 -0.0017641392 -0.0618098107 O -3.2591453480 -1.8898583454 -0.7264179834 Ti -5.1372070666 -1.8918692243 -0.0012386637 O -1.0878410276 -1.8908975100 -3.6681494905 Ti 2.2953566326 -1.8945802968 -0.7324918911 O 1.8572641703 -0.0016921856 -0.7344045642 O 4.0371444331 -1.8910414726 -1.5881377025 O 3.3210868792 -1.8907741455 0.8191885244 Ti -1.4878179598 -3.7811586802 -3.7059227721 O -1.0853532342 -3.7809408508 -1.6116052274 Ti -2.8450252555 -3.7788195849 -0.7274776599 O -3.3267379004 -3.7812381637 -2.7423527944 O -4.7300872942 -3.7790968457 -0.0598090260 O -3.2604741093 -5.6718710426 -0.7338292499 Ti -5.1447866872 -5.6729502955 -0.0741564861 O -1.0884608345 -5.6727912820 -3.6676316873 Ti 2.2684209803 -5.6717142765 -0.7217304029 O 1.8574125893 -3.7804564443 -0.7330818756 O 4.0285990372 -5.6721054737 -1.6077890803 O 3.3201243312 -5.6722897918 0.8087243331 Ti -1.4884820434 3.7822906244 -3.7073217712 O -1.0847762013 3.7818636329 -1.6113017246 Ti -2.8439453199 3.7841411650 -0.7265400935 O -3.3267270296 3.7819066970 -2.7433025606 O -4.7312054524 3.7823723470 -0.0697069103 O -3.2595288817 1.8897668415 -0.7334187674 Ti -5.1446141745 1.8933189943 -0.0754881482 O -1.0885182339 1.8906776867 -3.6679434552 Ti 2.2687722606 1.8941648243 -0.7213872473 O 1.8555780589 3.7824519637 -0.7335160805 O 4.0283551896 1.8906904137 -1.6083035670 O 3.3194888527 1.8901215845 0.8082989311 Ti 1.3702533850 -0.0107406935 3.4478828397 O -2.6375196242 -0.0002691731 -5.1384544425 Ti -4.2555253076 -0.0004346746 -4.2635860836 O -0.2706997157 -0.0070914550 4.3327078320 O 4.0252034756 -0.0000815787 -3.6655962284 O -4.7789243271 -1.8904283625 -4.5262265362 Ti 3.6276350884 -1.8913012322 -3.6941431294 O 1.8693336769 -1.8883429262 3.7243207414 Ti 0.8648978359 -1.8921733057 -4.2643698117 O 0.3356892769 -0.0005951329 -4.5283168344 O 2.4831990038 -1.8910320844 -5.1341360953 O 1.7891597814 -1.8911498456 -2.7417208102 Ti 1.3675607179 -3.7714249561 3.4584460387 O -2.6361368408 -3.7817733748 -5.1377491017 Ti -4.2537976019 -3.7816703567 -4.2621660812 O -0.2527486810 -3.7773208994 4.3296691681 O 4.0251551834 -3.7817133665 -3.6656274881 O -4.7792586102 -5.6728660382 -4.5273909859 Ti 3.6247553952 -5.6729206250 -3.7048960624 O 1.9006169442 -5.6721819263 3.7282320026 Ti 0.8602204280 -5.6728510136 -4.2616392365 O 0.3358111887 -3.7815283986 -4.5276229355 O 2.4785034911 -5.6729185089 -5.1373985723 O 1.7869078663 -5.6728807589 -2.7423620036 Ti 1.3817457800 3.7823928879 3.4589497419 O -2.6363646230 3.7820140473 -5.1385122329 Ti -4.2543090361 3.7822671573 -4.2626609244 O -0.2374835922 3.7781640807 4.3379058406 O 4.0248222240 3.7818879520 -3.6667895479 O -4.7794862202 1.8903770813 -4.5277962247 Ti 3.6243254290 1.8913422637 -3.7051707738 O 1.8974839341 1.8882474143 3.7289879155 Ti 0.8600377417 1.8920214690 -4.2620560393 O 0.3346421543 3.7821159876 -4.5275899679 O 2.4782941685 1.8909856222 -5.1376874872 O 1.7866749487 1.8910410218 -2.7427494459 H -3.2770955004 -2.4268873548 8.1687296457 C -3.1978647566 -1.3575831172 7.9359146369 H -4.2239524320 -0.9733256879 7.8414028480 H -2.6752907472 -0.8249390527 8.7355765151 C -2.5132448898 -1.1924834089 6.6141051521 O -2.8755597850 -1.8460118394 5.6163779603 O -1.5275649623 -0.3212592281 6.6009413157 H -1.0670534373 -0.2549619720 5.6840567874
- mode2.xyz
116 Ti 0.0150786099 -0.0030604155 -0.2374201259 O 0.4515580466 -0.0646282100 1.8001155831 Ti -1.5073356783 -0.0021026124 2.5525864818 O -1.7759961007 -0.0002275800 0.6680111337 O -3.3291162152 -0.0023490698 3.0696212715 O -1.7197127142 -1.8809866556 2.8642842575 Ti -3.6978004404 -1.8661667177 3.6084326925 O 0.3785074831 -1.8945596770 -0.1980298452 Ti 3.7996574045 -1.9219539316 2.8301270999 O 3.3305054330 -0.0058364403 2.7942261562 O -4.6900333011 -1.9659285303 1.9448925419 O -5.3925681292 -1.8976344334 4.3248895668 Ti -0.0283064667 -3.7737024590 -0.1507446117 O 0.4475870469 -3.7803762159 1.8621586431 Ti -1.3713031978 -3.7725215345 2.8511642577 O -1.7912822225 -3.7729870345 0.7436262925 O -3.1963784791 -3.7804631987 3.6648868231 O -1.7705984874 -5.6752809020 2.8039139185 Ti -3.7092032911 -5.6725438342 3.3897938268 O 0.3841910583 -5.6739517448 -0.1381224421 Ti 3.7513121067 -5.6747221635 2.8551954028 O 3.3367138059 -3.7840186471 2.8127851513 O -4.6560725160 -5.6733549533 1.8776383238 O -5.3204369054 -5.6669271369 4.2778026483 Ti -0.0266777619 3.7696389407 -0.1491534163 O 0.4524897086 3.7857220830 1.8631418152 Ti -1.3730627656 3.7742135603 2.8371003388 O -1.7905256319 3.7786525562 0.7422177730 O -3.1948630000 3.7795989881 3.6670918260 O -1.7343662633 1.8899845272 2.8090040137 Ti -3.6974632076 1.8668219133 3.5884163764 O 0.3822543310 1.8874843004 -0.1432095900 Ti 3.7864567490 1.9054527096 2.8508016360 O 3.3675002865 3.7799021368 2.8626057027 O -4.6840457398 1.9746044749 1.9466850498 O -5.3729095331 1.8414022655 4.3320391075 Ti -1.5079668291 -0.0008456767 -3.7989215478 O -1.0951317039 0.0022938033 -1.7345576883 Ti -2.8929121223 -0.0000735048 -0.8782772455 O -3.3736661811 0.0005357865 -2.8128806833 O -4.7193863680 0.0001570116 -0.0748056115 O -3.2632860440 -1.8860154449 -0.7956216654 Ti -5.1307996411 -1.8978086035 -0.0398179095 O -1.1320787396 -1.8909803808 -3.7440163178 Ti 2.3133857413 -1.9061341614 -0.8099170994 O 1.8786839357 0.0011314062 -0.8069543100 O 4.0385009963 -1.8895616393 -1.6570045786 O 3.3115055560 -1.8920970172 0.7501369741 Ti -1.5256127358 -3.7819843403 -3.7701763163 O -1.0940683369 -3.7869847697 -1.6789541127 Ti -2.8425313294 -3.7819322313 -0.7839726892 O -3.3422315516 -3.7760564219 -2.8030699366 O -4.7315665522 -3.7831312336 -0.1321164185 O -3.2580399316 -5.6728032840 -0.7985618810 Ti -5.1462151180 -5.6726106968 -0.1400092932 O -1.1340144859 -5.6732616791 -3.7388880843 Ti 2.2670101177 -5.6749205566 -0.7762329495 O 1.8631140907 -3.7853073574 -0.7974118985 O 4.0263575475 -5.6730177766 -1.6715945206 O 3.3250517248 -5.6784227751 0.7515944134 Ti -1.5248867442 3.7818848512 -3.7712820230 O -1.0907299886 3.7812336768 -1.6778258037 Ti -2.8428177895 3.7841675976 -0.7861032158 O -3.3397902383 3.7762483047 -2.8026235400 O -4.7304156316 3.7842200412 -0.1318958639 O -3.2645051173 1.8869138058 -0.7933488063 Ti -5.1241443302 1.8993249071 -0.0252297449 O -1.1314283131 1.8906347021 -3.7440419862 Ti 2.3185671036 1.9017475834 -0.7833845676 O 1.8597419566 3.7800089897 -0.7974665372 O 4.0344536794 1.8895028649 -1.6315658326 O 3.3098310076 1.8936248995 0.7823280807 Ti 1.4537853090 -0.0347286638 3.2604875735 O -2.6845024000 0.0000746592 -5.2147510734 Ti -4.2967571219 0.0018586454 -4.3442818967 O -0.2511143909 0.1328085915 4.2720687411 O 3.9819696258 0.0012627538 -3.7355131655 O -4.8038476957 -1.8889916108 -4.5923792314 Ti 3.5953547314 -1.8907810205 -3.7642624098 O 1.8553660493 -1.8815919187 3.6062633048 Ti 0.8351449362 -1.8959349617 -4.3196742869 O 0.3119042183 -0.0001283684 -4.5915870901 O 2.4377532123 -1.8915585194 -5.2027330310 O 1.7783437087 -1.8888222418 -2.8082665928 Ti 1.3904951140 -3.7857957693 3.3690175525 O -2.6814201117 -3.7831847959 -5.2025558657 Ti -4.2854691947 -3.7801054852 -4.3136778334 O -0.2074321278 -3.7970194791 4.2709539045 O 3.9820256895 -3.7800854351 -3.7356644708 O -4.7961377238 -5.6736624399 -4.5471769110 Ti 3.5917963887 -5.6730897244 -3.7666434973 O 1.9069112439 -5.6781980199 3.6136248160 Ti 0.8312484616 -5.6732029738 -4.3044789530 O 0.3186267326 -3.7822693739 -4.5479829736 O 2.4335197920 -5.6733247069 -5.1986406627 O 1.7786689446 -5.6734212363 -2.7982114552 Ti 1.3984968329 3.7644681343 3.3685503422 O -2.6815455972 3.7828337111 -5.2024703087 Ti -4.2855071694 3.7788568387 -4.3128750844 O -0.2044150611 3.7319133608 4.2704391005 O 3.9827213914 3.7789077586 -3.7330838039 O -4.8039829955 1.8898269577 -4.5907748518 Ti 3.5899147654 1.8912350407 -3.7590482617 O 1.8739898095 1.8720733323 3.5992207033 Ti 0.8344557677 1.8954291350 -4.3129140724 O 0.3192643376 3.7819866970 -4.5466934222 O 2.4382493211 1.8919495801 -5.1986096411 O 1.7788752457 1.8885973165 -2.8025129463 H -1.3211320733 -0.5901398003 7.3622691450 C -2.2774669737 -0.0487846563 7.3616031246 H -2.9713818519 -0.6195053843 7.9928436643 H -2.1415146457 0.9598101494 7.7628564958 C -2.8081585120 -0.0061954447 5.9550273922 O -3.0662415287 -1.1345711119 5.4074245732 O -2.9500441444 1.1398319329 5.4010622063 H -0.3897742635 1.0153182782 4.6757696096