exercises:2017_ethz_mmm:qmmm
Table of Contents
Validation of a KCl QMMM model
(exercise by Matthew Watkins, University college, London)
In this exercise you will validate the mixed quamtum/classical model for a KCl slab. The present exercise is referring to the following paper: 10.1002/jcc.23904.
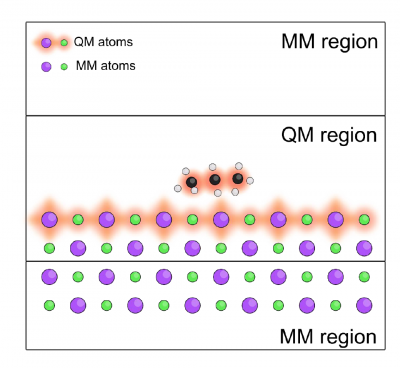
- You will optimize the geometry of a KCL slab with the same arrangement as the one depicted below
- For simplicity, you will consider three layers.
- You will compare the calculation performed with the full QM, one layer QM and two MM, two layer QM and one MM.
- In particular you will compare the band gaps and the density of states.
1. Task: Prepare the input files
The file input.inp contains the partition of the QM and MM regions by specifying the atom index. Check in the kcl.xyz how the indexes are distributed. The slab orthogonal coordinate is the y.
- Make three copies of input.inp and call them qm_1l.inp, qm_2l.inp and qm.inp.
- qm_1l.inp should have
PROJECT KCl_1and one QM layer. Modify accordingly the MM_INDEX lines by looking first at thekcl.xyzcoordinate files. - qm_2l.inp should have
PROJECT KCl_2and two QM layers. Modify accordingly the MM_INDEX lines by looking first at thekcl.xyzcoordinate files. - qm.inp should have
PROJECT KCl_QMand the full QM treatment. For this, it is sufficient to changeQMMMat the beginning toQS. Change also the input coordinates from kcl.xyz to kcl_opt.xyz: these are the already optimized coordinates for the full QM treatment. In this way you will spare time.
2. Task: Run the jobs
- Run the jobs by giving the command:
qsub run -v INP=qm_1land similarly for the other input files. - You will also get cube files for hartree potential and electronic density. They can be examined with vmd.
- For each job, a
*pos*xyzoptimization file is produced, as well as two PDOS files, one for the species 1 (K), the other for the species 2 (Cl). All will be prefixed by thePROJECTprefix you set in task 1.
3. Task: Checking the geometry
By direct inspection of the last configuration in each file
*pos*xyz (example: KCl_QM-pos-1.xyz), check the distance between 1-2, 2-3 layers. What are the differences between the three cases?
4. Task: Electronic properties
Extract a smeared dos from the
pdos files, using the python scripts present in the directory. For each case in task 1 the procedure is:
> python get-smearing-pdos.py KCl_1-PDOS-QMMM-k1-1.pdos > mv smeared.dat KCl_1.K.dat > python get-smearing-pdos.py KCl_1-PDOS-QMMM-k2-1.pdos > mv smeared.dat KCl_1.Cl.dat > paste KCl_1.K.dat KCl_1.Cl.dat > KCl_1.KCl.dat # all in the same file. The columns are ''ENERGY PDOS_K ENERGY PDOS_Cl ''
Plot the .dat files using gnuplot. The Fermi energy is set to zero eV.
> gnuplot gnuplot> set xrange [-5:10] gnuplot> plot 'KCl_1.KCl.dat' u 1:($2+$4) w l, plot 'KCl_QM.KCl.dat' u 1:($2+$4) w l, plot 'KCl_2.KCl.dat' u 1:($2+$4) w l
- Note the differences you observe
- Find the value of the band gap in the
*.outfiles and relate it to what you see in gnuplot. How close is the QMMM to the full QM representation?
Required Files
When you are dealing with complex job structure, the input can be simplified by splitting it into multiple files. We are going to use separate files for the coordinates, the QM part, the MM part. All these files should reside in the same directory as the main input file.
The provided files are all in the directory
/home/psd/Exercise_13. When you create new input files with different parameters, remember to change the name of the PROJECT as well.
- input.inp
@SET METHOD = QMMM # FIST all classical treatment # QS all quantum treatment &GLOBAL FLUSH_SHOULD_FLUSH PRINT_LEVEL LOW PROJECT KCl RUN_TYPE GEO_OPT &END GLOBAL &FORCE_EVAL METHOD $METHOD @include QS.inc @include MM.inc &QMMM #this defines the QS cell in the QMMM calc &CELL ABC 12.6 15.0 12.6 PERIODIC XZ &END CELL ECOUPL GAUSS # use GEEP method NOCOMPATIBILITY USE_GEEP_LIB 6 # use GEEP method &PERIODIC # apply periodic potential #in this case QM box = MM box in XZ so turn #off coupling/recoupling of the QM multipole &MULTIPOLE OFF &END &END PERIODIC #these are just the ionic radii of K Cl #but should be treated as parameters in general #fit to some physical property &MM_KIND Cl RADIUS 1.67 &END MM_KIND #define the model &QM_KIND K MM_INDEX 25..32 41..48 &END QM_KIND &QM_KIND Cl MM_INDEX 17..24 33..40 &END QM_KIND &END QMMM &SUBSYS #this defines the cell of the whole system #must be orthorhombic, I think &CELL ABC 12.6 100.0 12.6 &END CELL &TOPOLOGY COORD_FILE_NAME kcl.xyz COORD_FILE_FORMAT XYZ &GENERATE &ISOLATED_ATOMS #ignores bonds dihedrals etc in classical part LIST 1..48 &END &END &END &KIND K ELEMENT K BASIS_SET DZVP-MOLOPT-SR-GTH POTENTIAL GTH-PBE-q9 &END KIND &KIND Cl BASIS_SET DZVP-MOLOPT-GTH POTENTIAL GTH-PBE-q7 &END &END SUBSYS &END FORCE_EVAL #should be able to use most motion sections #analytic stress tensor not available, I think @include motion.inc
and includes as separate files, using the @include macro, for the QS, MM and motion sections
- QS.inc
&DFT BASIS_SET_FILE_NAME BASIS_MOLOPT POTENTIAL_FILE_NAME GTH_POTENTIALS &MGRID COMMENSURATE CUTOFF 150 &END MGRID &QS EPS_DEFAULT 1.0E-12 &END QS &SCF EPS_SCF 1.0E-06 MAX_SCF 26 SCF_GUESS RESTART &OT MINIMIZER CG PRECONDITIONER FULL_SINGLE_INVERSE ENERGY_GAP 0.001 &END OT &OUTER_SCF EPS_SCF 1.0E-05 &END OUTER_SCF &END SCF &XC &XC_FUNCTIONAL PBE &END XC_FUNCTIONAL &END XC &PRINT &MO_CUBES NLUMO 10 WRITE_CUBE T &END MO_CUBES &V_HARTREE_CUBE STRIDE 2 2 2 &END &END PRINT &END DFT
- MM.inc
&MM &FORCEFIELD &CHARGE ATOM K CHARGE 1.0 &END CHARGE &CHARGE ATOM Cl CHARGE -1.0 &END CHARGE &NONBONDED &WILLIAMS atoms K Cl A [eV] 4117.9 B [angstrom^-1] 3.2808 C [eV*angstrom^6] 0.0 RCUT [angstrom] 3.0 &END WILLIAMS &WILLIAMS atoms Cl Cl A [eV] 1227.2 B [angstrom^-1] 3.1114 C [eV*angstrom^6] 124.0 RCUT [angstrom] 3.0 &END WILLIAMS &WILLIAMS atoms K K A [eV] 3796.9 B [angstrom^-1] 3.84172 C [eV*angstrom^6] 124.0 RCUT [angstrom] 3.0 &END WILLIAMS &END NONBONDED &END FORCEFIELD &POISSON &EWALD EWALD_TYPE spme ALPHA .44 GMAX 40 &END EWALD &END POISSON &END MM
- motion.inc
&MOTION &GEO_OPT OPTIMIZER LBFGS &END &CONSTRAINT &FIXED_ATOMS LIST 1..16 EXCLUDE_MM .FALSE. EXCLUDE_QM .TRUE. &END FIXED_ATOMS &END CONSTRAINT &END MOTION
- kcl.xyz
48 Cl 0.00000 15.00000 0.00000 Cl 3.15000 15.00000 3.15000 Cl 0.00000 15.00000 6.30000 Cl 3.15000 15.00000 9.45000 Cl 6.30000 15.00000 0.00000 Cl 9.45000 15.00000 3.15000 Cl 6.30000 15.00000 6.30000 Cl 9.45000 15.00000 9.45000 K 3.15000 15.00000 0.00000 K 0.00000 15.00000 3.15000 K 3.15000 15.00000 6.30000 K 0.00000 15.00000 9.45000 K 9.45000 15.00000 0.00000 K 6.30000 15.00000 3.15000 K 9.45000 15.00000 6.30000 K 6.30000 15.00000 9.45000 Cl 3.15000 18.15000 0.00000 Cl 0.00000 18.15000 3.15000 Cl 3.15000 18.15000 6.30000 Cl 0.00000 18.15000 9.45000 Cl 9.45000 18.15000 0.00000 Cl 6.30000 18.15000 3.15000 Cl 9.45000 18.15000 6.30000 Cl 6.30000 18.15000 9.45000 K 0.00000 18.15000 0.00000 K 3.15000 18.15000 3.15000 K 0.00000 18.15000 6.30000 K 3.15000 18.15000 9.45000 K 6.30000 18.15000 0.00000 K 9.45000 18.15000 3.15000 K 6.30000 18.15000 6.30000 K 9.45000 18.15000 9.45000 Cl 0.00000 21.30000 0.00000 Cl 3.15000 21.30000 3.15000 Cl 0.00000 21.30000 6.30000 Cl 3.15000 21.30000 9.45000 Cl 6.30000 21.30000 0.00000 Cl 9.45000 21.30000 3.15000 Cl 6.30000 21.30000 6.30000 Cl 9.45000 21.30000 9.45000 K 3.15000 21.30000 0.00000 K 0.00000 21.30000 3.15000 K 3.15000 21.30000 6.30000 K 0.00000 21.30000 9.45000 K 9.45000 21.30000 0.00000 K 6.30000 21.30000 3.15000 K 9.45000 21.30000 6.30000 K 6.30000 21.30000 9.45000
exercises/2017_ethz_mmm/qmmm.txt · Last modified: 2020/08/21 10:15 by 127.0.0.1