This is an old revision of the document!
Table of Contents
Protein Folding in Solution
In this exercise, you will calculate the protein folding free energy in aqueous solution using thermodynamic integration, a method based on molecular dynamics (MD). The protein will be described by the empirical force field, CHARMM, http://mackerell.umaryland.edu/charmm_ff.shtml
Background
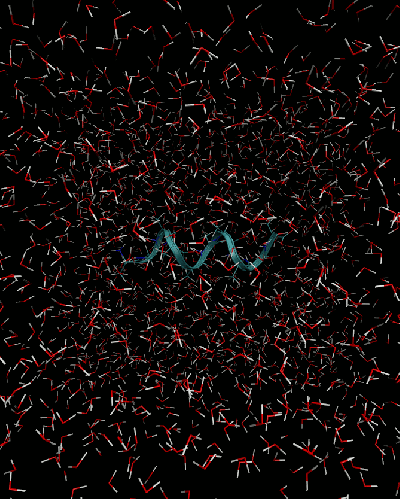
A model protein you will have to deal with is the alanine decapeptide. The folding/unfolding will be achieved by fixing the distance between the end carbon atoms in the chain: atoms 7 and 98. This distance is called a collective variable. At each distance one runs the MD simulation (constrained MD) to extract the time-averaged forces acting on the collective variable, $F(x)$. Then, a free energy difference can be calculated via thermodynamic integration (TI):
\begin{equation} \Delta A = -\int_a^b F(x)\, dx \end{equation}
Here $a$ and $b$ are the initial and the final values of the collective variable. TI is a general method, which can be applied to a variety of processes, e.g. phase transitions, electron transfer etc.
Task 1: Familiarize yourself
Download the files: deca_ala.tar.gz
deca_ala.pdb (protein data base) file contains the coordinates
deca_ala.psf (protein structure file) file contains connectivity data
par_all27_prot_lipid.inp contains the force field parameters
md_1836.inp is the CP2K input file
Open the deca_ala.pdb protein data bank format file with vmd. Create a new representation for the protein, e.g. of type Ribbon to observe the alpha-helix.
Task 2: Perform constrained MD simulations
For that you have to run MD for different values of the distance between atoms 7 and 98, in each run it will be constrained. In the original file md_1836.inp it is set to $18.36$ Å as is in the deca_ala.pdb file.
- Run CP2K with
md_1836.inp - Copy
md_1836.inpto smth. likemd_1536.inp; - Modify the PROJECT_NAME and
TARGETvalue in theCONSTRAINTsection for a new value: here 15.36; - Run CP2K with the new input file;
- Repeat for several values in the range $15$ to $20 $ Å.
- To avoid confusion, try to perfrom every task in a new directory
- You may increase or decrease the number of MD steps, which is set to 5000 in the file, to speed-up the calculation or else get a better statiscics.
Constraint section TO BE modified for constrained MD
- constraint section
&CONSTRAINT &COLLECTIVE COLVAR 1 INTERMOLECULAR TARGET [angstrom] 18.36 &END COLLECTIVE &LAGRANGE_MULTIPLIERS COMMON_ITERATION_LEVELS 1 &END &END CONSTRAINT
Task 3: Evaluate the free energy difference
⇒ Each constrained MD will produce a .LagrangeMultLog-files, which look like this:
Shake Lagrangian Multipliers: -63.547262596 Rattle Lagrangian Multipliers: 63.240598387 Shake Lagrangian Multipliers: -0.326901815 Rattle Lagrangian Multipliers: -0.318145579
- From these files you can calculate the average Lagrange multiplier of the Shake-algorithm like this:
grep Shake yourprojectname.LagrangeMultLog | awk '{c++ ; s=s+$4}END{print s/c}'
- The average Lagrange multiplier is the average force $F(x)$ required to constrain the atoms at the distance $x$.
- From these forces the free energy difference can be obtained via TI (see Background)
