This is an old revision of the document!
Dehydration of ethanol
We will investigate today a very important chemical reaction, the production of ethene (ethylene) from ethanol. Ethanol is heated with an excess of concentrated sulphuric acid at a temperature of 170°C. The gases produced are passed through sodium hydroxide solution to remove the carbon dioxide and sulphur dioxide produced from side reactions. Another way to favor this reaction is in presence of a catalyst surface, which makes the reaction exothermic even at room temperature, and the free energy barrier for the reaction is highly reduced, so to obtain reasonable reaction rates. The use of hematite (iron-based nanocrystal) as a catalyst is described computationally in the following paper by Lopes et al: 10.1039/c6ra08509a. We are interested in the gas phase reaction, in particular in the reactants and the products energy, to estimate the “energy of reaction”.
The reaction is the following:
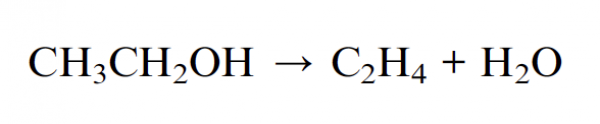 and this is the pictorial view in the gas phase:
and this is the pictorial view in the gas phase:
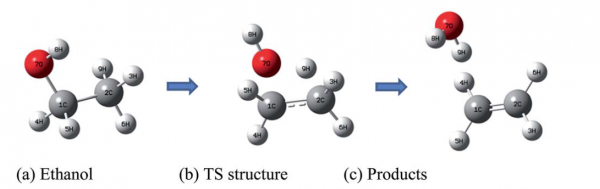
In the supplementary material of the paper, which can be found here, several calculation with different methods ranging from DFT to hybrid methods to high-level methods are described, together with the basis set dependence of the results.
We will compare our results with the published ones.
The input file structure is shown below:
&FORCE_EVAL ! This section defines method for calculating energy and forces
METHOD Quickstep ! Electronic structure method (DFT,...)
&DFT
BASIS_SET_FILE_NAME ./BASIS_C_H_O
&POISSON ! Solver requested for non periodic calculations
PERIODIC NONE
PSOLVER WAVELET ! Type of solver
&END POISSON
&QS ! Parameters needed to set up the Quickstep framework
METHOD GAPW ! Method: gaussian and augmented plane waves
&END QS
#
# Include the exchange and correlation information
#
@INCLUDE './hf.inc'
&END DFT
&SUBSYS ! This section defines the system
&CELL ! Unit cell set up
PERIODIC NONE ! Non periodic calculation
ABC 10 10 10 ! Lengths of the cell vectors A, B, and C
&END CELL
&COORD ! This section specify all the atoms and their coordinates
H 2.5558925119 3.0 4.
O 4.00 3.5 4.
H 5.4167134504 3.0669045998 4.
&END COORD
&TOPOLOGY
&CENTER_COORDINATES
&END
&END TOPOLOGY
&KIND O ! potential and basis for O
BASIS_SET _MYBASIS_
POTENTIAL ALL
&POTENTIAL
4 4 0
0.24762086 0
&END
&END KIND
&KIND H ! potential and basis for H
BASIS_SET _MYBASIS_
POTENTIAL ALL
&POTENTIAL
1 0 0
0.20000000 0
&END
&END KIND
&KIND C ! potential and basis for C
BASIS_SET _MYBASIS_
POTENTIAL ALL
&POTENTIAL
4 2 0
0.34883045 0
&END
&END KIND
&END SUBSYS
&END FORCE_EVAL ! This section defines method for calculating energy and forces
&GLOBAL ! Section with general information regarding which kind of simulation to perform an parameters for the whole PROGRAM
PRINT_LEVEL LOW ! Global print level
PROJECT h2o ! Name of the project. This word will appear as part of a name of all ouput files (except main ouput file, specified with -o option)
RUN_TYPE GEO_OPT ! Geometry optimization
&END GLOBAL
The exercise is executed on the virtual machine cluster. After creating a directory as usual, you can download the file from here: (tar file)
Copy the file to the created directory in the EXERCISES path, and execute
tar xvf exercise_5.tar
h2o_template.inpfor the geometry optimization of waterethanol_template.inpfor the geometry optimization of ethanolethylene_template.inpfor the geometry optimization of ethylene (ethene)hf.incfile describing the level of theory (exchange and/or correlation functional), in this case Hartree FockBASIS_H_C_Oa repository file for basis sets.
First you have to modify the input files in order to prescribe a certain basis set. You have, from the simplest to the most advanced:
- 3-21G*
- 6-31G*
- 6-31Gxx
- 6-31++Gxx
- 6-311Gxx
- 6-311++Gxx
For example,
m_replace _MYBASIS_ '6-31G*' < h2o_template.inp > h2o_BASIS1.inp
(note: use the quote, because the star is a bash special character!!!) The command to launch the job is
In the *.out files you can look for the final energies by the following command:
.
This will list all the energies. Then you can redo the exercise with different basis set, reusing the command m_replace.
| Basis set | Energy H2O [$E_h$] | Energy Ethylene [$E_h$] | Energy Ethanol [$E_h$] | Distance CC Ethanol [$Å$] | Reaction energy [$E_h$] |
|---|---|---|---|---|---|
| BASIS 1(Name) | …. | …. | …. | …. | …. |
| BASIS 2(Name) | …. | …. | …. | …. | …. |
| BASIS 3(Name) | …. | …. | …. | …. | …. |
| BASIS 4(Name) | …. | …. | …. | …. | …. |
| BASIS 5(Name) | …. | …. | …. | …. | …. |
| BASIS 6(Name) | …. | …. | …. | …. | …. |
- Compute the reaction energy for the dehydration reaction of ethanol
- Prepare a table with rows and columns: on the rows the basis set, on the columns different quantities (see above).
- Compare the results with the published ones (note the conversion factors. You can use the tool at Energy converter
- Comment on the dependence on the basis set
- Is this information enough to determine the rates of reaction? Why?
BONUS TRACK
ethanol_BASIS1.inp into ethanol_dens_BASIS1.inp.
Add the following sections: under &DFT
&PRINT
&E_DENSITY_CUBE
&END
&END
&SCF
SCF_GUESS RESTART
&END
This tells to read the old wavefunction and to print the cubefile of the density.
At the end of the input file:
&EXT_RESTART
RESTART_FILE_NAME ./c2h6o-1.restart
&END
Then, change RUN_TYPE GEO_OPT to RUN_TYPE ENERGY to only run a single point calculation. It will generate a cubefile with the density which you may visualize with VMD.
