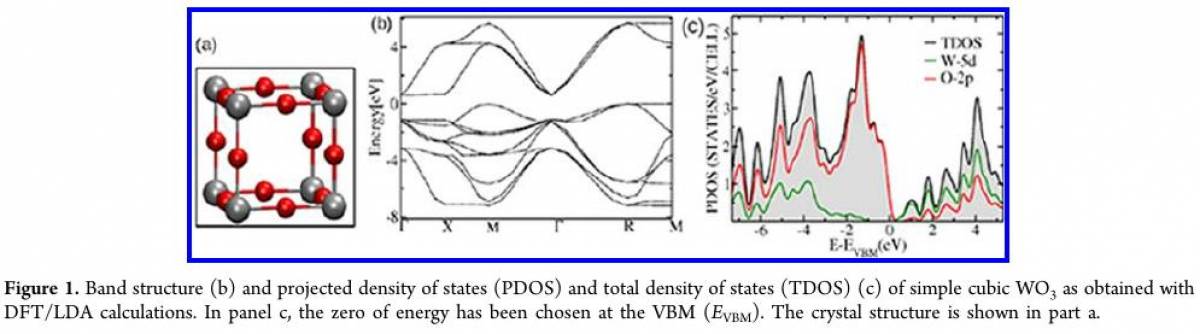
Getting the band structure of WO$_3$ Lattice
In this exercise, you will carry out band structure calculation using K-point sampling for Cubic lattice WO$_3$. The reference band structure you can find in this paper
To get the band structure for WO3, only a few changes are required compared to the previous example for calculating the PDOS:
- WO3-bs.inp
&GLOBAL PROJECT WO3-kp-bs RUN_TYPE ENERGY PRINT_LEVEL MEDIUM &END GLOBAL &FORCE_EVAL METHOD Quickstep &DFT BASIS_SET_FILE_NAME BASIS_MOLOPT POTENTIAL_FILE_NAME POTENTIAL &POISSON PERIODIC XYZ &END POISSON &QS EXTRAPOLATION USE_GUESS ! required for K-Point sampling &END QS &SCF SCF_GUESS ATOMIC EPS_SCF 1.0E-6 MAX_SCF 300 ADDED_MOS 2 &DIAGONALIZATION ALGORITHM STANDARD EPS_ADAPT 0.01 &END DIAGONALIZATION &SMEAR ON METHOD FERMI_DIRAC ELECTRONIC_TEMPERATURE [K] 300 &END SMEAR &MIXING METHOD BROYDEN_MIXING ALPHA 0.2 BETA 1.5 NBROYDEN 8 &END MIXING &END SCF &XC &XC_FUNCTIONAL PBE &END XC_FUNCTIONAL &END XC &KPOINTS SCHEME MONKHORST-PACK 3 3 1 SYMMETRY OFF WAVEFUNCTIONS REAL FULL_GRID .TRUE. PARALLEL_GROUP_SIZE 0 &END KPOINTS &PRINT &BAND_STRUCTURE ADDED_MOS 2 FILE_NAME WO3.bs &KPOINT_SET UNITS B_VECTOR SPECIAL_POINT ??? #GAMA SPECIAL_POINT ??? #X SPECIAL_POINT ??? #M SPECIAL_POINT ??? #GAMA SPECIAL_POINT ??? #R SPECIAL_POINT ??? #M NPOINTS ??? &END &END BAND_STRUCTURE &END PRINT &END DFT &SUBSYS &CELL ABC [angstrom] 3.810000 3.810000 3.810000 PERIODIC XYZ MULTIPLE_UNIT_CELL 1 1 1 &END CELL &TOPOLOGY MULTIPLE_UNIT_CELL 1 1 1 &END TOPOLOGY &COORD SCALED W 0.0 0.0 0.0 O 0.5 0.0 0.0 O 0.0 0.5 0.0 O 0.0 0.0 0.5 &END &KIND W ELEMENT W BASIS_SET DZVP-MOLOPT-SR-GTH POTENTIAL GTH-PBE &END KIND &KIND O ELEMENT O BASIS_SET DZVP-MOLOPT-SR-GTH POTENTIAL GTH-PBE &END KIND &END SUBSYS &END FORCE_EVAL
Some notes on the input file:
- By specifying the
KPOINTsection you are enabling the K-Point calculation. - While you could specify the K-Points directly, we are using the Monkhorst-Pack scheme 1) to generate them. The numbers following the keyword
MONKHORST-PACKspecify the tiling of the brillouin zone. - After the basic calculation, CP2K calculates the energies along certain lines, denoted as
KPOINT_SET(when you check the documentation you will note that this section can be repeated). - The keyword
NPOINTSspecifies how many points (in the addition to the starting point) should be sampled between two special points. - The
SPECIAL_POINTkeyword is used to specify the start-, mid- and endpoints of the line. Those points usually denote special points in the reciprocal lattice/unit cell, like the $\Gamma$, $M$ or $K$ point. You can find the definition for these in the appendix section of this paper. This keyword can also be specified multiple times, making it possible to directly get the band structure for a complete path.
Now, when you run this input file you will get in addition the the output file, a file named WO3.bs which will look similar to the following:
SET: 1 TOTAL POINTS: 26
POINT 1 ******** ******** ********
POINT 2 ******** ******** ********
POINT 3 ******** ******** ********
POINT 4 ******** ******** ********
POINT 5 ******** ******** ********
POINT 6 ******** ******** ********
Nr. 1 Spin 1 K-Point 0.00000000 0.00000000 0.00000000
20
-73.66652408 -38.53370023 -37.80464132 -37.79327769
-16.71308703 -16.11075946 -16.02553853 -1.43495530
-1.34739188 -1.33357408 0.37912017 0.38948689
0.39582882 0.40030859 0.46965212 0.47418816
2.60728842 2.62105342 3.16044140 6.99806305
Nr. 2 Spin 1 K-Point 0.00000000 0.10000000 0.00000000
20
-73.66647294 -38.53337818 -37.80859042 -37.79536623
-16.67479677 -16.09554462 -15.96731960 -1.68492873
-1.44087258 -1.34318045 0.09257368 0.13769271
0.21643888 0.38447849 0.44179002 0.45757924
2.61768501 3.02368022 3.51828287 7.06644645
[...]
For each set there is a block named SET with the special points listed as POINT, followed by sub-blocks for each K-Point containing the energies for each MO.
Your tasks:
- Lookup the special points for the $\Gamma$, $X$,$M$,$R$ points in the mentioned paper (make sure you choose the right lattice). Calculate and plot the band structure for WO3 lattice along $\Gamma$, $X$,$M$,$\Gamma$,$R$,$M$ (you are free to decide whether to use multiple K-Point sets are multiple special points in a single set). Mark the special points. Choose an appropriate number of interpolation points to get a smooth plot.
- Compare your plot with plots from literature. What is different?
- How many orbital energies do you get and why? Try to change the input to get more unoccupied orbitals.
To convert the band structure file to a file which can be plotted directly, you can use the script cp2k_bs2csv.py from below, which when passed a band structure file WO3.bs as an argument will write files WO3.bs-set-1.csv for each set containing the K-Point coordinates and the energies in one line.
To plot the WO3.bs-set-1.csv file, you can either load it into $MATLAB$ or use $GNUPLOT$ command line.
gnuplot>set yrange [-8:14] gnuplot>plot for [i=4:23] "WO3.bs.set-1.csv" u 0:i w l t ""
- cp2k_bs2csv.py
#!/usr/bin/env python """ Convert the CP2K band structure output to CSV files """ import re import argparse SET_MATCH = re.compile(r''' [ ]* SET: [ ]* (?P<setnr>\d+) [ ]* TOTAL [ ] POINTS: [ ]* (?P<totalpoints>\d+) [ ]* \n (?P<content> [\s\S]*?(?=\n.*?[ ] SET|$) # match everything until next 'SET' or EOL ) ''', re.VERBOSE) SPOINTS_MATCH = re.compile(r''' [ ]* POINT [ ]+ (?P<pointnr>\d+) [ ]+ (?P<a>\S+) [ ]+ (?P<b>\S+) [ ]+ (?P<c>\S+) ''', re.VERBOSE) POINTS_MATCH = re.compile(r''' [ ]* Nr\. [ ]+ (?P<nr>\d+) [ ]+ Spin [ ]+ (?P<spin>\d+) [ ]+ K-Point [ ]+ (?P<a>\S+) [ ]+ (?P<b>\S+) [ ]+ (?P<c>\S+) [ ]* \n [ ]* (?P<npoints>\d+) [ ]* \n (?P<values> [\s\S]*?(?=\n.*?[ ] Nr|$) # match everything until next 'Nr.' or EOL ) ''', re.VERBOSE) if __name__ == '__main__': parser = argparse.ArgumentParser(description=__doc__) parser.add_argument('bsfilename', metavar='bandstructure-file', type=str, help="the band structure file generated by CP2K") args = parser.parse_args() with open(args.bsfilename, 'r') as fhandle: for kpoint_set in SET_MATCH.finditer(fhandle.read()): filename = "{}.set-{}.csv".format(args.bsfilename, kpoint_set.group('setnr')) set_content = kpoint_set.group('content') with open(filename, 'w') as csvout: print(("writing point set {}" " (total number of k-points: {totalpoints})" .format(filename, **kpoint_set.groupdict()))) print(" with the following special points:") for point in SPOINTS_MATCH.finditer(set_content): print(" {pointnr}: {a}/{b}/{c}".format( **point.groupdict())) for point in POINTS_MATCH.finditer(set_content): results = point.groupdict() results['values'] = " ".join(results['values'].split()) csvout.write("{a} {b} {c} {values}\n".format(**results))