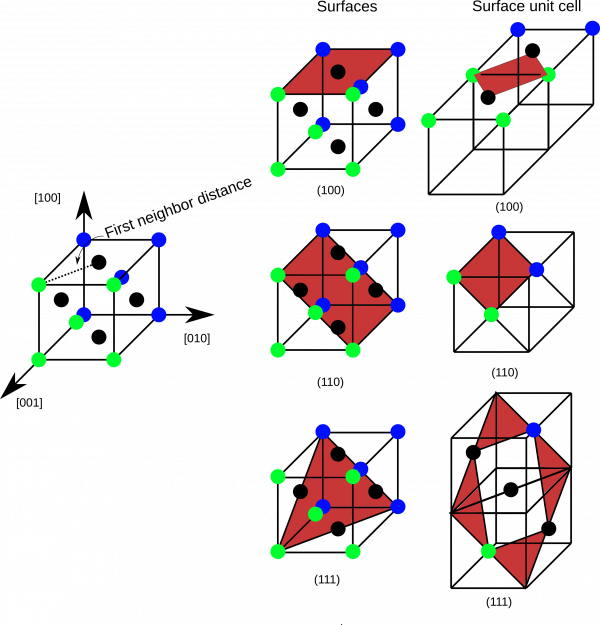
Surface energies of Copper high-symmetry surfaces
you@eulerX ~$ module load courses mmm vmd
you@eulerX ~$ mmm-init
you@eulerX ~$ module load new cp2k
and to submit the job:
you@eulerX ~$ bsub < jobname
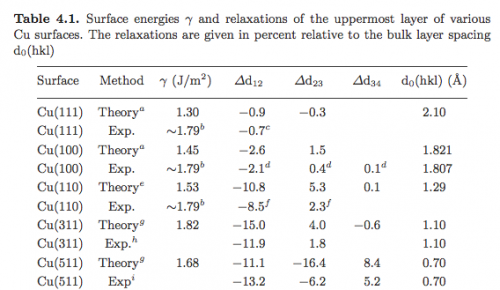
In this exercise we will compute the surface energies of Cu using the EAM potential. As a reference, we report the table from the Gross book:
- Download all the necessary files from from the wiki: e4.1.zip (all inputs are commented) in your home directory and unzip it:
you@eulerX ~$ wget http://www.cp2k.org/_media/exercises:2017_ethz_mmm:e4.1.zip you@eulerX ~$ unzip exercises:2017_ethz_mmm:e4.1.zip you@eulerX ~$ cd exercise_4.1
If you cannot download with wget, you can copy the zip file directly from my directory
you@eulerX ~$ cp /cluster/home/danielep/2017/e4.1.zip . you@eulerX ~$ unzip e4.1.zip you@eulerX ~$ cd exercise_4.1
- Run the optimizations 100.inp, 110.inp, 111.inp and the bulk.
you@eulerX exercise_4.1$ bsub cp2k.popt -i 100.inp -o 100.out you@eulerX exercise_4.1$ bsub cp2k.popt -i 110.inp -o 110.out you@eulerX exercise_4.1$ bsub cp2k.popt -i 111.inp -o 111.out you@eulerX exercise_4.1$ bsub cp2k.popt -i bulk.inp -o bulk.out
- While geometry optimization is running you can have a look at the corresponding initial coordinate files 100.xyz, 110.xyz, 111.xyz. In vmd it is also possible to open a console, and give the command pbc set { a b c 90 90 90 } where a, b, c can be extracted from the cp2k input file. Then you can make several periodic copies for visualization.
you@eulerX exercise_4.1$ vmd 100.xyz you@eulerX exercise_4.1$ vmd 110.xyz you@eulerX exercise_4.1$ vmd 111.xyz
- Without calculating the surface energies but just looking at the structures, could you guess which surface will be the most stable one? And the least stable one? Why? (Quite often a simple logic could tell you already how your results should look like.)
- Have a look into the files, containing the optimization trajectory (100-pos-1.xyz, 110-pos-1.xyz, 111-pos-1.xyz). Along the optimization procedure all the surface structures make a similar movement. Could you describe it and explain why it is happening? Take into account that all the geometries were taking from the bulk structure.
- Compute the three surface energies: you need to compute the area, subtract bulk contribution, take care of the units.
Now we will learn how to compute the Wulff crystal from these three numbers. Basically, you should edit the input file “DP”, replacing “PUT_HERE_THE_XXX_SURFACE_ENERGY” with corresponding surface energy. Note that for the {100} and {110} surfaces you need to edit TWO lines each: the program takes care of assigning the surface energy to equivalent surfaces (like (010)) but since it was developed for island on surfaces it does not do it automatically in all directions. z direction is treated differently in this program.
- At this point, you can run the sowos program:
you@eulerX exercise_4.1$ ./sowos.v02.00.02.x
- There will be many output files. Important are:
- the atomistic model out.atomistic-inside-gnuplot.xyz which will contain many atoms if you chose a proportionality constant in the file DP which is too large. Unfortunately this file does NOT have a proper structure of an xyz file.
- This can be solved by using a simple m_* function that adds the atomic symbol at the beginning of each line, and adding two lines at the beginning of the resulting file (number of atoms; comment):
you@eulerX exercise_4.1$ m_addcolumn Au < out.atomistic-inside-gnuplot.xyz > cluster.xyz you@eulerX exercise_4.1$ nano cluster.xyz # Add two lines at the beginning.
- the out.plot-gnuplot.plt file. You can open it with gnuplot
you@eulerX exercise_4.1$ gnuplot gnuplot> load "out.plot-gnuplot.plt" gnuplot> set xrange [-80:80] gnuplot> set yrange [-80:80] gnuplot> set view equal xyz gnuplot> replot
you can rotate with the mouse!
- What is the proper structure of a XYZ file? Try to appropriately modify out.atomistic-inside-gnuplot.xyz and open it in vmd. Make a nice snapshot and add it to your report.
- Use the cluster generated with SOWOS as an input configuration for a cluster optimization with cp2k. Comment on the final geometry. BEWARE OF THE CELL! It is not a periodic system!